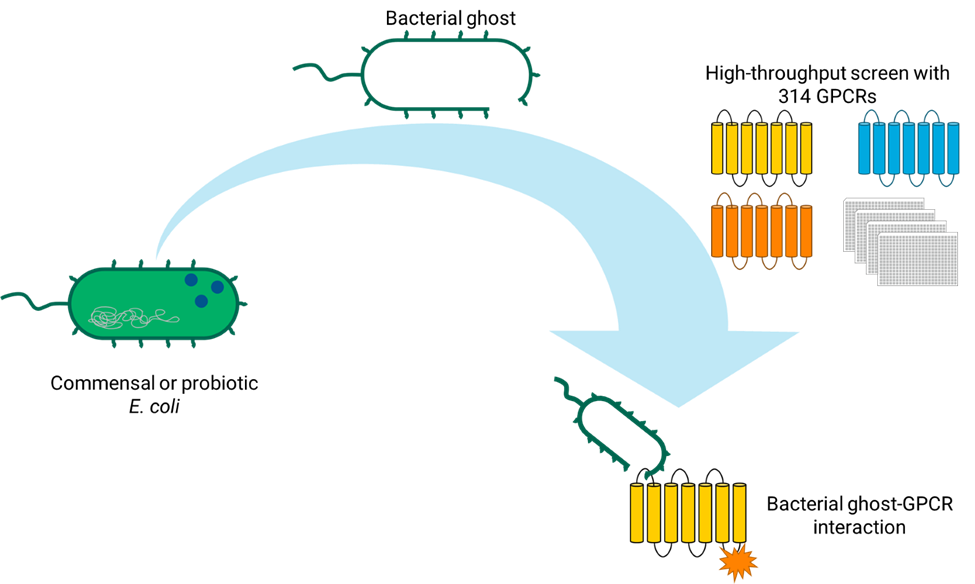
Bacteria display surface components that can interact with receptors found within the body. These surface components are known to interact with G-protein coupled receptors (GPCRs), a superfamily of transmembrane receptors that are often overexpressed in certain disease conditions. Interactions between bacterial surface components and GPCRs can facilitate bacterial adherence and internalization or can regulate an immune response by hijacking receptor signaling. We hypothesize that by exploiting these bacteria-GPCR interactions, we can develop effective targeted drug delivery carriers. However, the extent that these interactions occur remain unclear since conventional methods only investigate one receptor-ligand pair at a time. Our approach is to produce bacterial ghosts that will allow us to use high-throughput screening techniques to explore interactions between bacterial surface components and hundreds of GPCRs. Ghost bacteria are non-living bacterial cells devoid of their internal components with a fully intact membrane. This project ultimately aims to use these bacterial ghosts as a drug delivery vehicle targeting GPCRs. Bacterial ghosts from two commensal E. coli strains and a probiotic E. coli strain were chemically produced using dilute concentrations of a cell lysis buffer. Interactions between these ghost bacteria and 314 GPCRs were then explored using a high-throughput screening technique that measures GPCR signaling. Successful bacterial ghost production was confirmed by measuring DNA release and imaging the intact membrane with SEM. A preliminary high-throughput screening with these bacterial ghosts identified interactions that overlap with known bacteria-host pathways and identified previously unexplored interactions. Future work is dedicated to validating initial hits, orthogonal assays to probe the functional response of these interactions, and drug loading/release studies. Overall, this study presents a platform to produce bacterial ghosts and to explore their interactions in a high-throughput manner, allowing us to perform the first high-throughput screen between whole bacteria and hundreds of GPCRs.