Affiliation: Distinguished Professor, Purdue University; Martha & Fred Borch Chair in Cancer Therapeutics; Associate Dean for Graduate Programs, College of Pharmacy; Purdue Center for Cancer Research
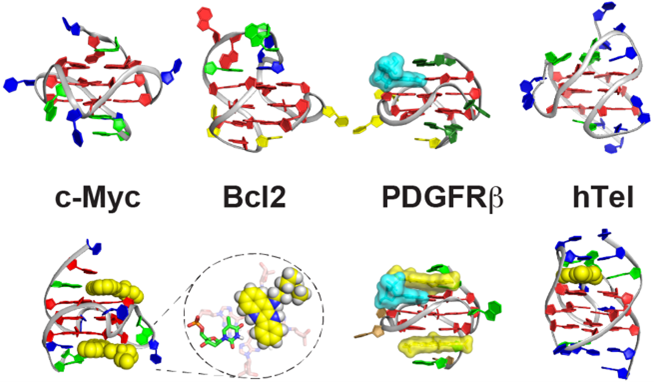
Abstract: DNA G-quadruplexes are secondary structures formed in guanine-rich regions of oncogene promoters and human telomeres. These structures are functionally significant and represent a promising class of cancer-specific drug targets. In this talk, I will present our efforts to elucidate the structures of G-quadruplexes in key oncogene promoters (MYC, BCL2, PDGFR-β, VEGF, KRAS) and human telomeres, and their interactions with small molecules, using high-field NMR spectroscopy. I will also discuss our progress in developing G4-targeted anticancer therapeutics, with a particular focus on MYC inhibitors.
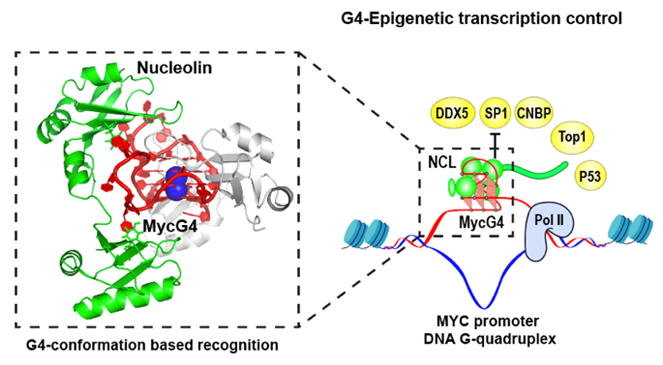
I will also present our recent work on protein interactions with the MYC oncogene promoter G-quadruplex, a transcriptional silencer. We discovered that the DEAD-box helicase DDX5 binds strongly to the MYC G-quadruplex and actively unwinds it to promote MYC transcription. On the other hand, nucleolin functions as a transcriptional repressor by binding and stabilizing the MYC G-quadruplex. We determined the 2.6 Å crystal structure of the nucleolin–MYC G-quadruplex complex, representing the first example of G4-conformation-based protein recognition. The structure reveals that nucleolin achieves high-affinity and specific binding through multivalent interactions involving its four RNA-binding domains. These findings underscore the role of G-quadruplexes as epigenetic regulators that modulate transcription in concert with interacting proteins and highlight their potential as molecular targets for MYC-focused cancer therapeutics.
Reference:
1. Chen L, Dickerhoff J, Sakai S, Yang, D. (2022) Accounts of Chemical Research. 55 (18), 2628 https://doi.org/10.1021/acs.accounts.2c00337
2. Wu G, Xing Z, Tran EJ, Yang D. (2019) PNAS, 116 (41), 20453. Doi:10.1073/pnas.1909047116.
3. Chen, Yang, et al., (2025) Science, 388(6744). https://doi.org/10.1126/science.adr1752